ScMaSigPro R-package
ScMaSigPro is an R package designed for analyzing Single-cell RNA-Seq Trajectory Data. It uses polynomial GLMs to model differential gene expression along Pseudotime, effectively helping you analyze single-cell trajectory branches.

Pathway Metagene Activity
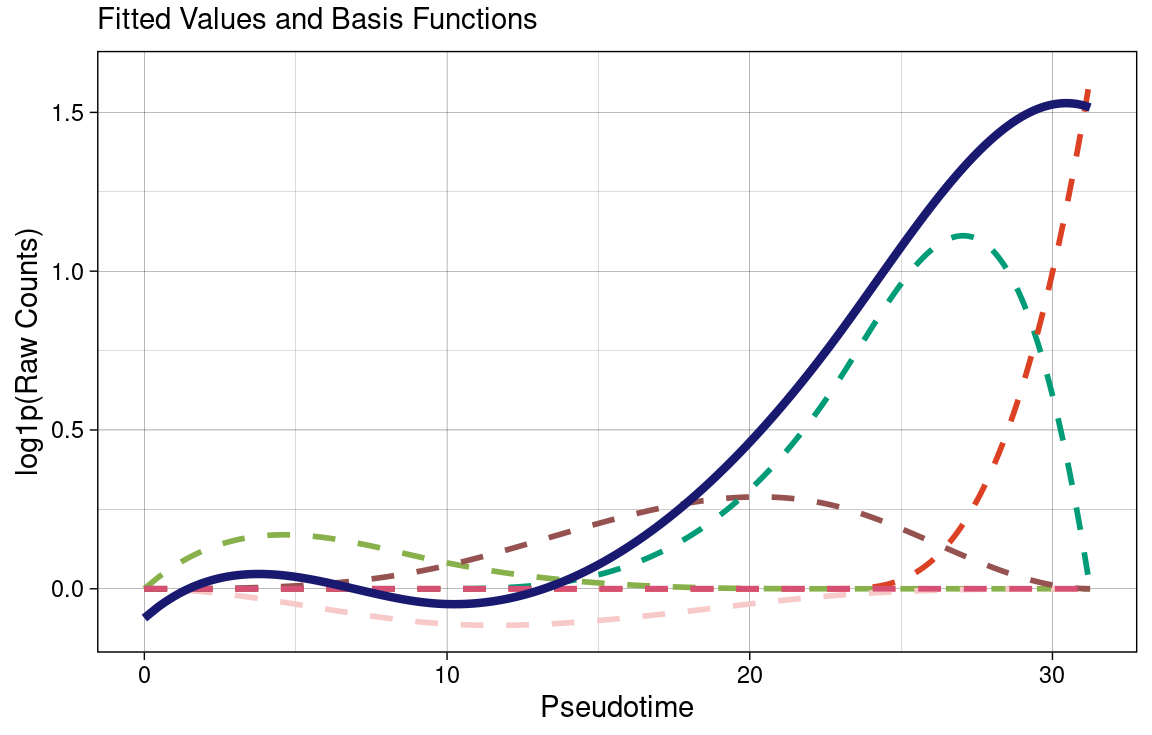
To put it simply, genes do not function in isolation; they interact with each other and form pathways. The notion of a Metagene attempts to explain these behaviors using matrix factorization. This project aims to explain these interactions in terms of Pseudotime. I am currently working on this project.
Nextflow Script for Metagenomic Analysis
(Master’s Project) This project aimed to evaluate whether clustering microbiome gene sequences at varying levels of sequence similarity (97% to 100%) influences the inferred microbial interactions in correlation networks. Specifically, it assess the relative importance of habitat-driven microbial associations under these clustering strategies. The pipleine is evaluted remotely using nextflow (each process in a conda environment) and the results are visualized using R. The ‘Assign Taxanomy’ process is ran within a EC2-Instance due to memory limitations.
CUT&RUN Data Analysis
Snakemake Pipeline to analyze data produced using Cleavage Under Targets and Release Using Nuclease protocol sequenced on Illumina NovaSeq 6000.
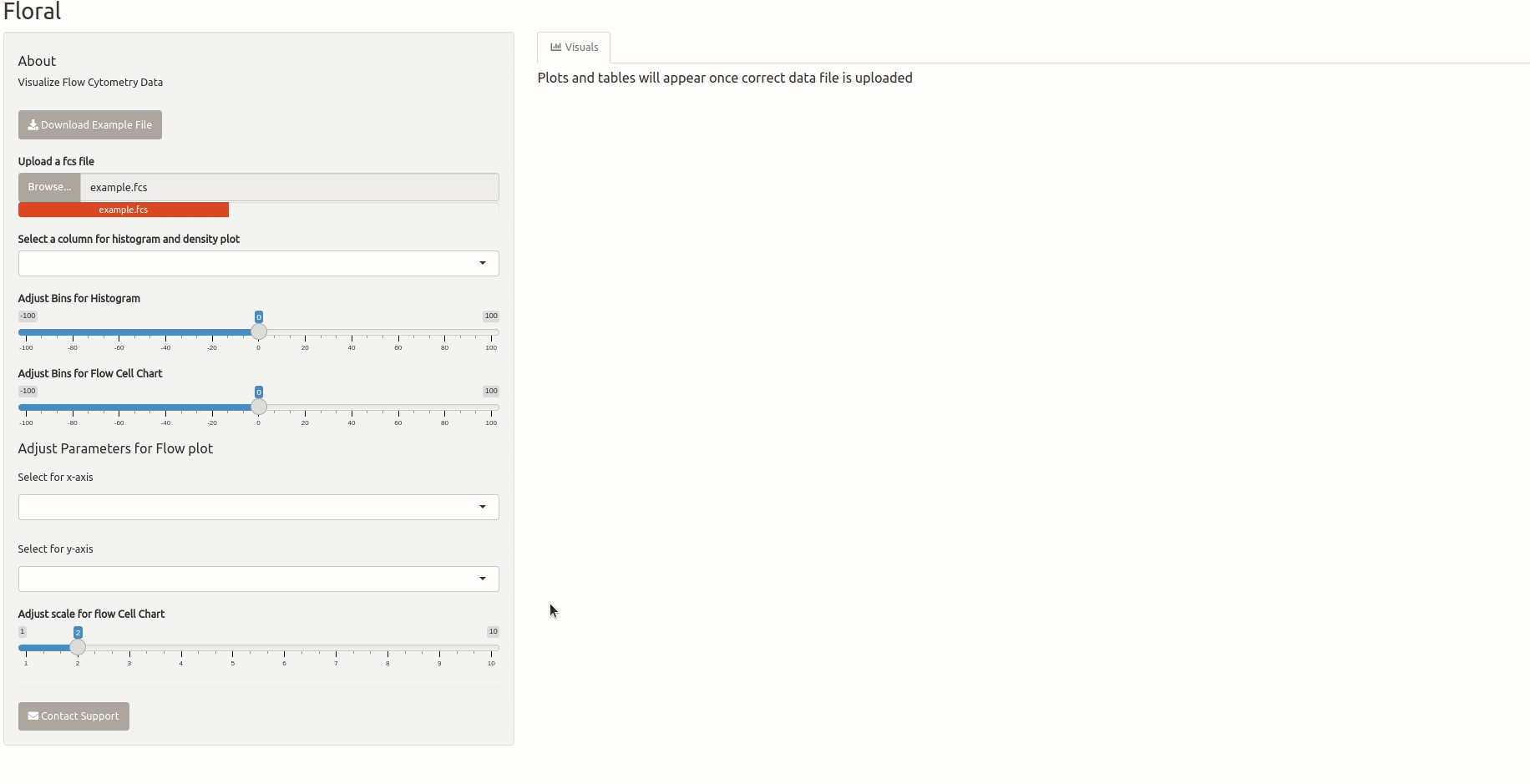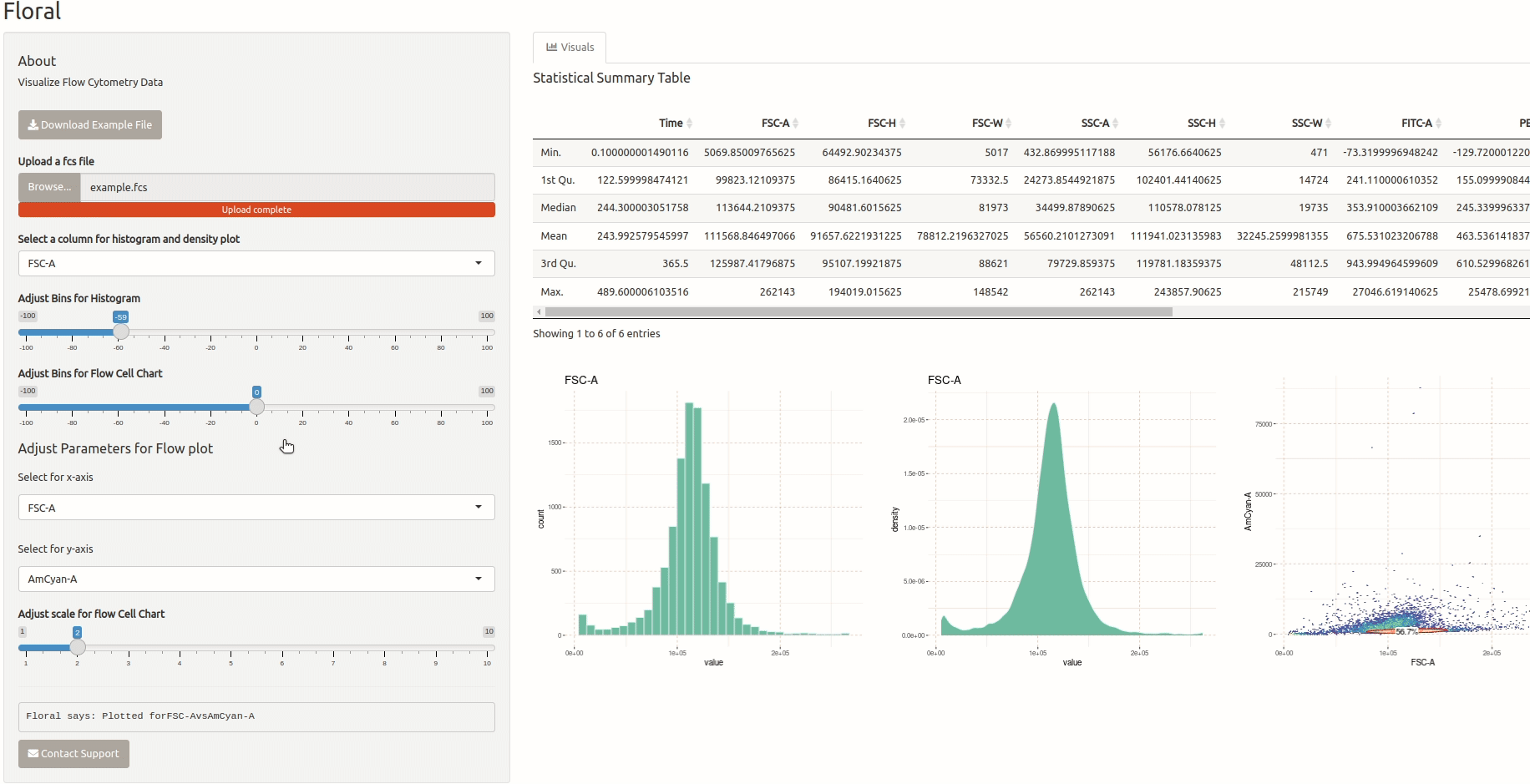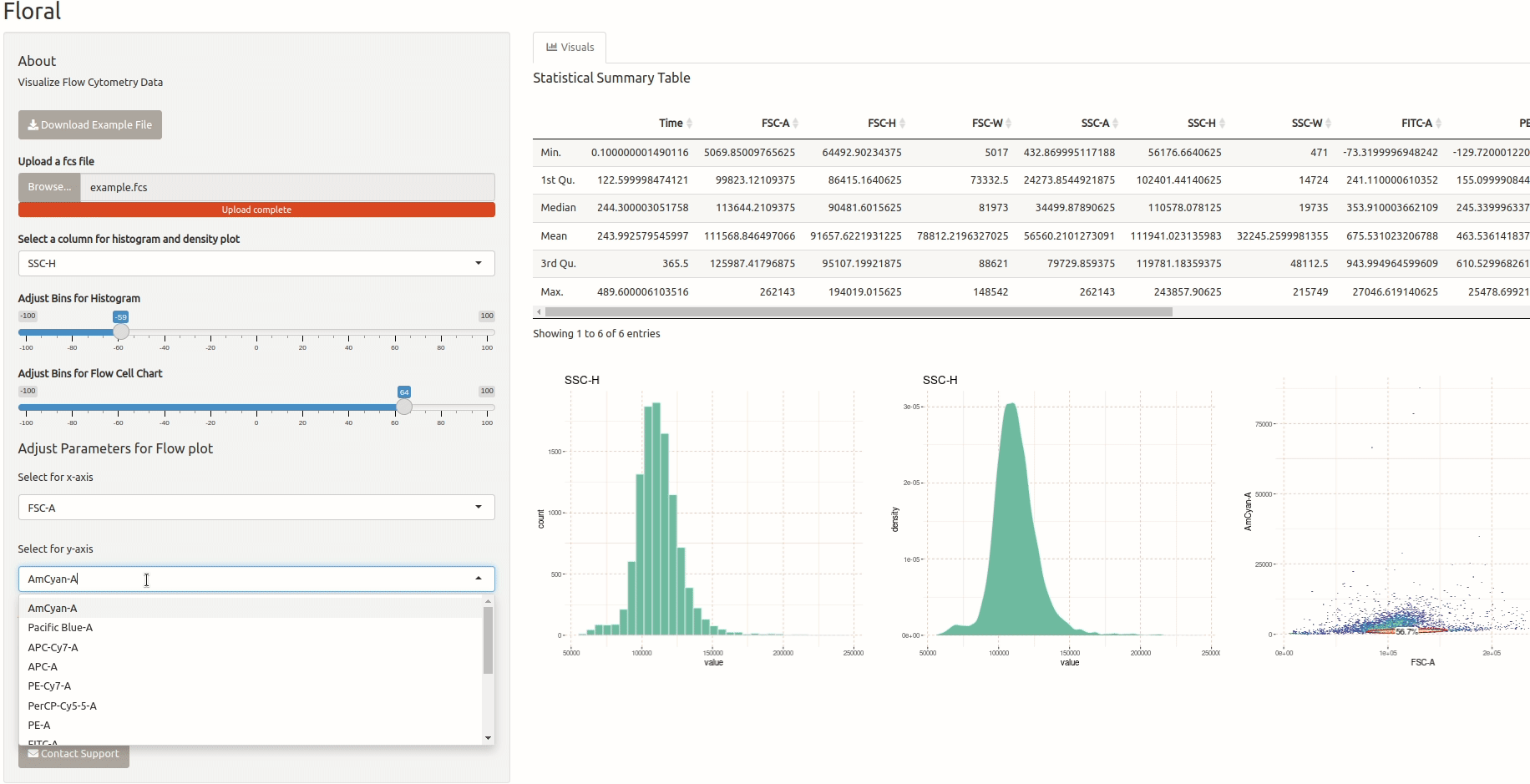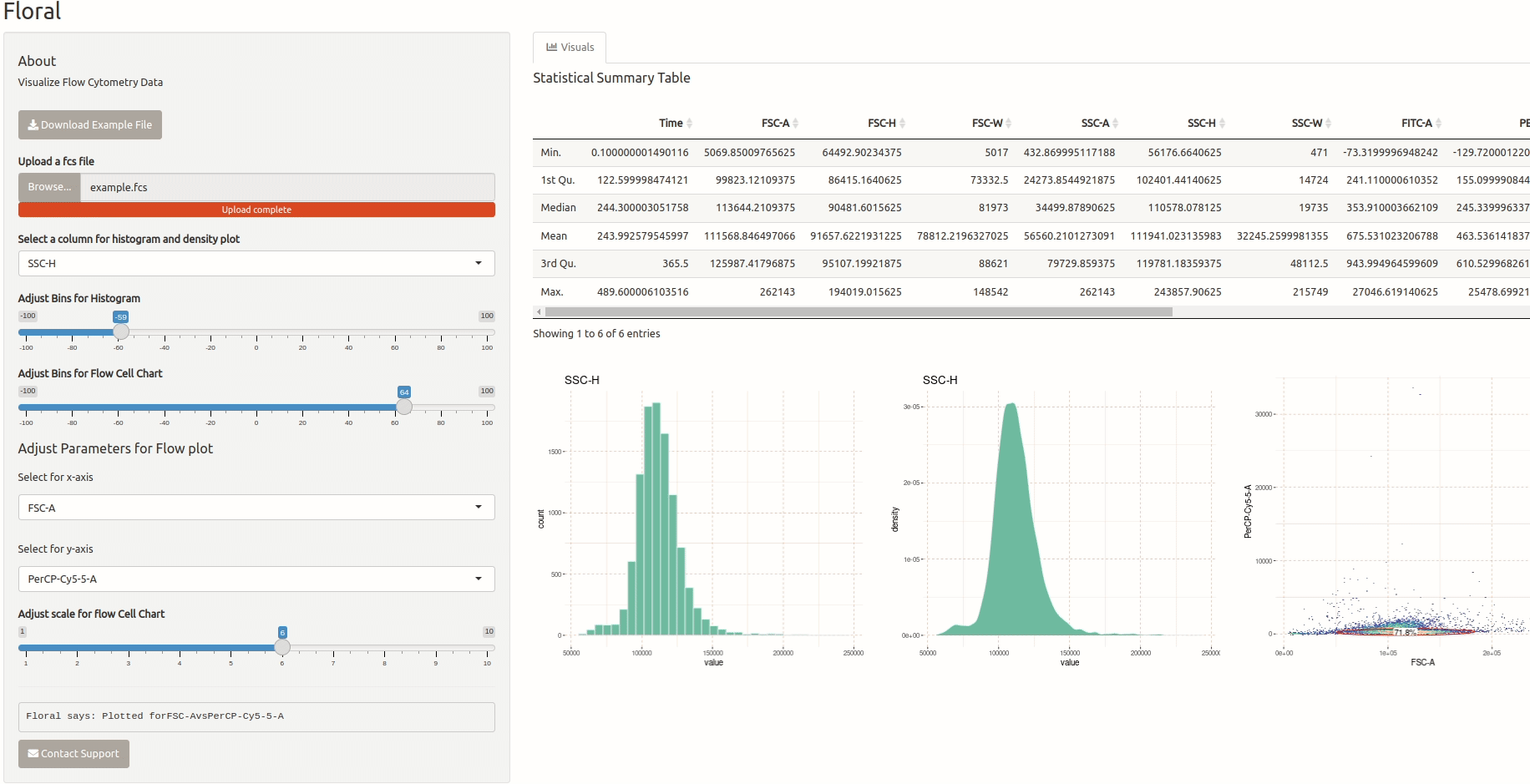
Visualization of Flow Cytometry Data
This is a small app that allows users to upload an FCS file containing flow cytometry data and visualize it. The app utilizes the ‘flowCore’ package to read the FCS file and the ‘ggcyto’ package to plot the data. Users can also select specific channels they want to visualize. I developed this app for a friend who is a biologist and needed a tool to visualize flow cytometry data.
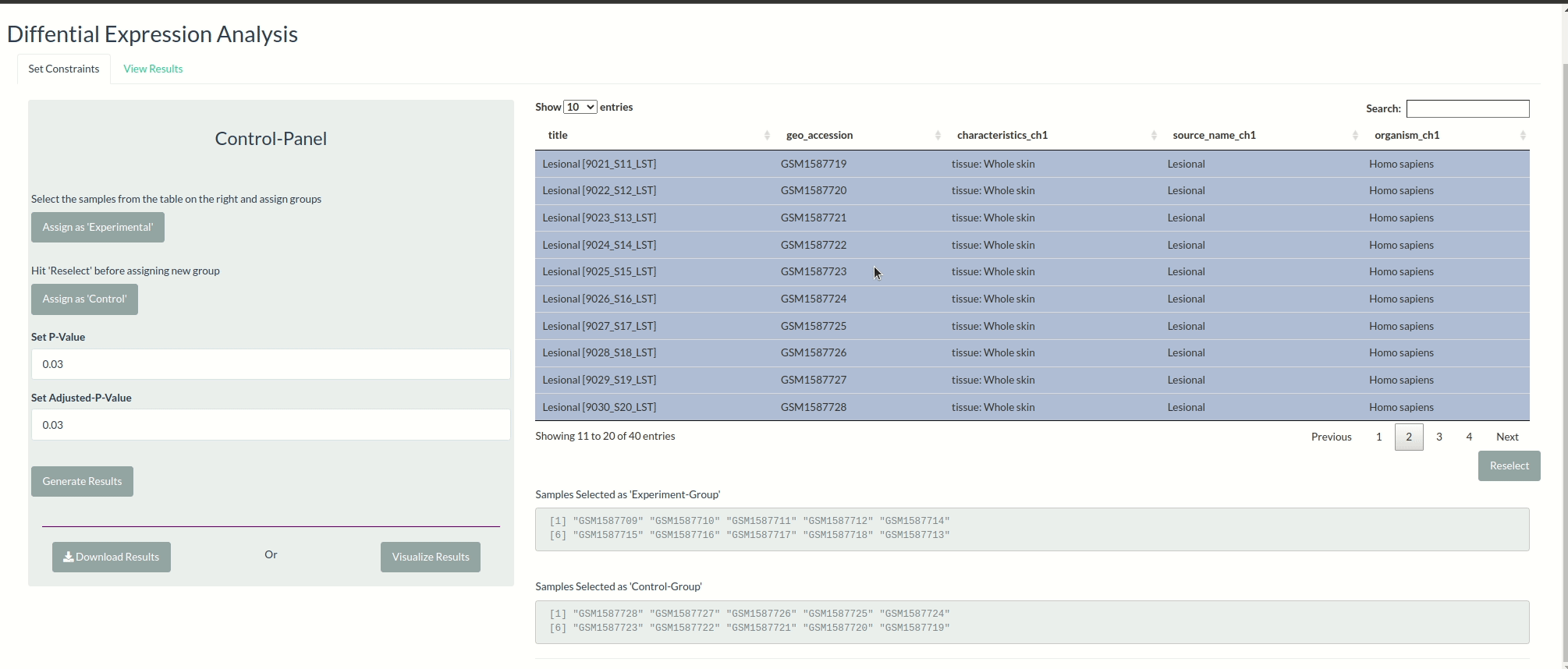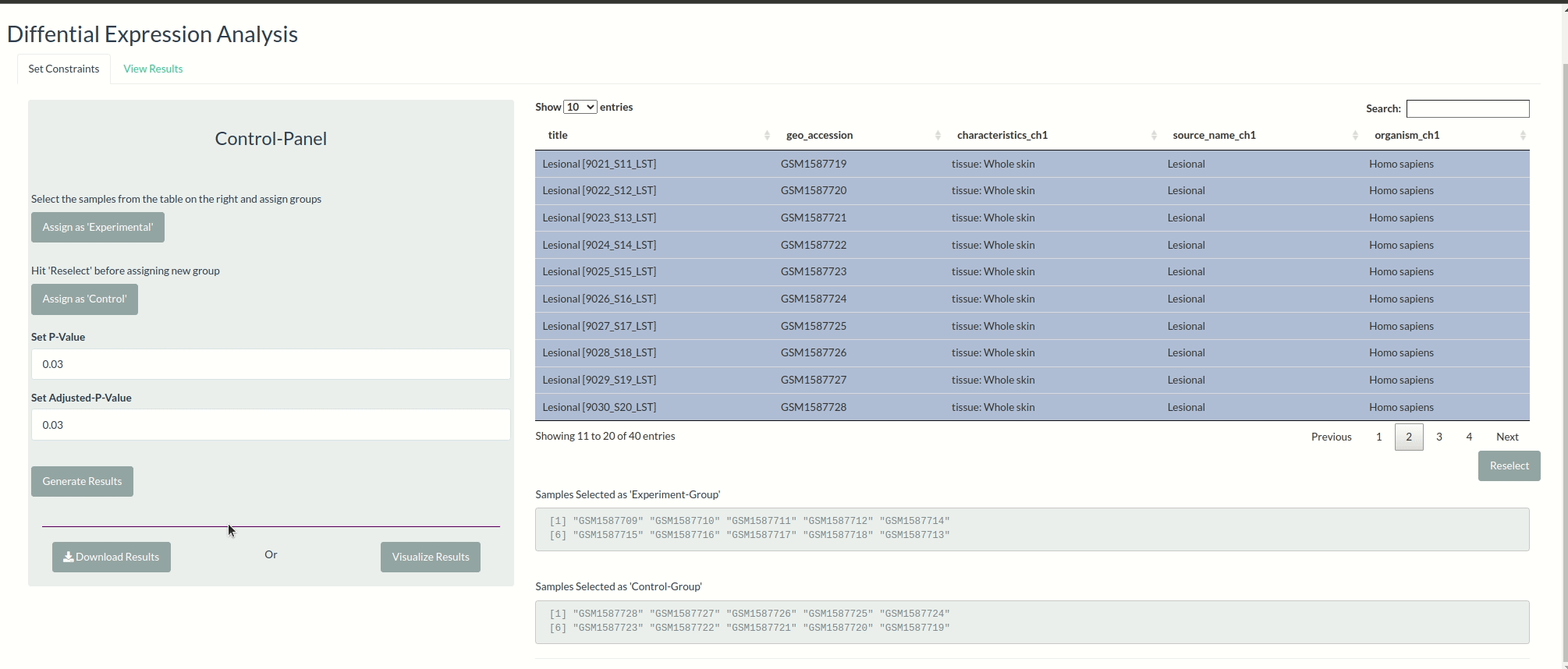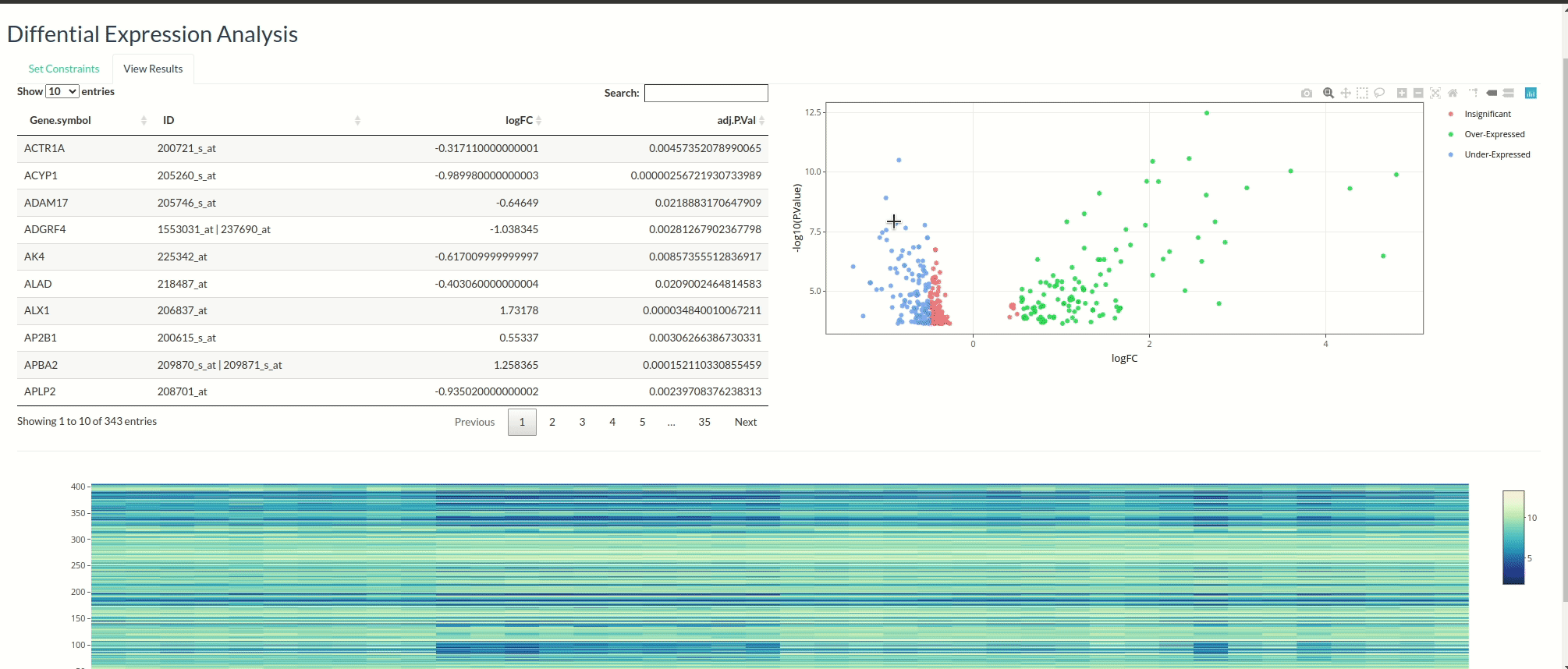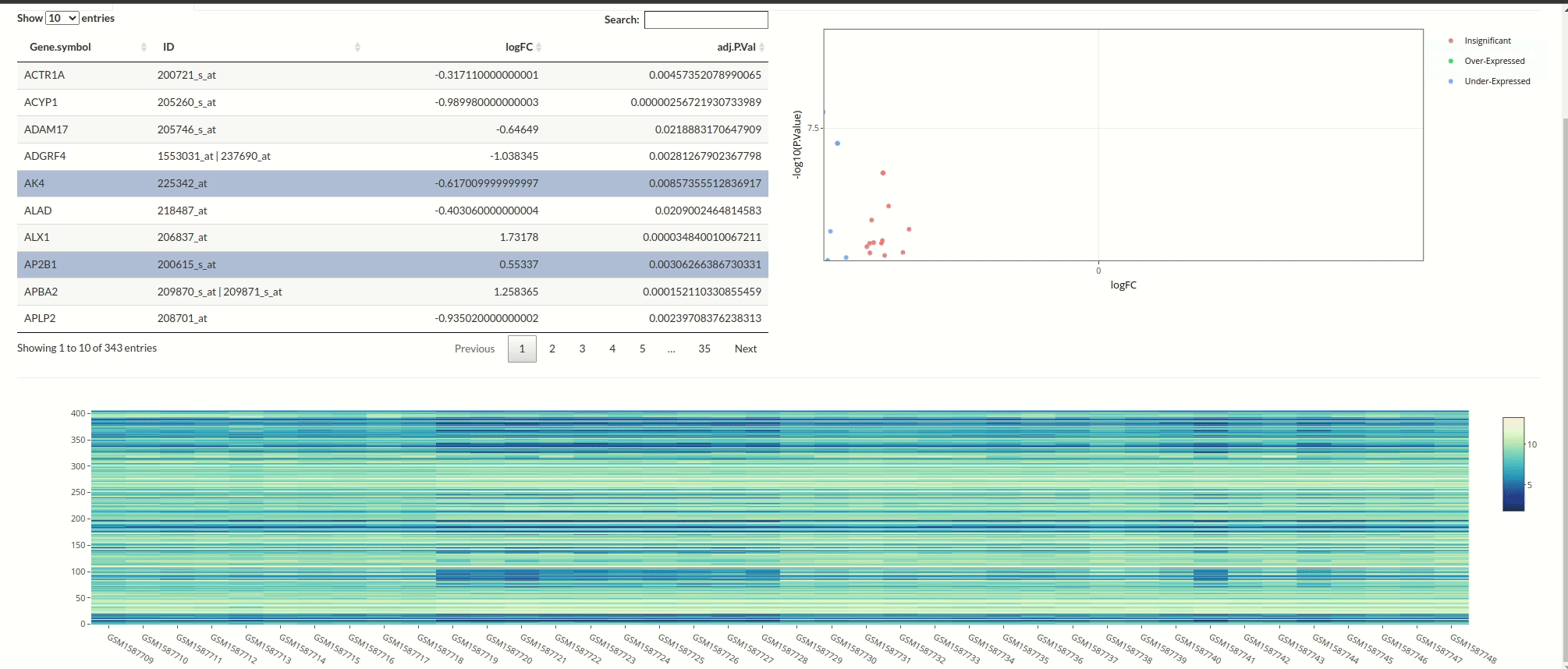
Differential Expression Analysis with Limma
This was my very first project using Shiny. The app allows users to directly fetch datasets from the GEO database and interactively select samples. I chose not to focus too much on the visual design since it was my first project. To begin the analysis, start with the ‘Fetch’ tab!
More?
Back in 2018, the first development framework I learned was R-Shiny. Since then, I have been a huge fan of its simplicity and flexibility. Over the years, I have developed several dashboards for various projects. Initially, I published them on shinyapps.io, but now I am migrating them to AWS-EC2.
I’m still crafting this website and updating this page. Stay tuned for more!